Entities¶
Class IRIs¶
Standard OBO purl, see page on how to set up Protege.
All classes are in MONDO ID space
Class Metadata¶
The standard OBO properties are used:
- Label. Every class MUST have a single unambiguous label
- Definition. Every class SHOULD have a text definition.
- Synonyms. Use broad/narrow/exact/related wisely.
- See: uberon synonyms guide.
- TODO: still to clean up a lot of synonym scopes seeded from external ontologies
- We tend to use BROAD/NARROW generously, even if the sub/super exists. This is because it is useful to annotate other ontologies usages of synonyms.
- Comment: Optional. max one per class (obo-format limit).
- Use of more specific annotation properties is recommended. These are being hacked into comments at the moment. (e.g Editor note)
Labels¶
Case rules:
- Use lowercase, even for initial letter, except for these exceptions:
- proper names (for example, Epstein-Barr virus-associated mesenchymal tumor)
- latin names (for example, Homo sapiens)
- acronyms (e.g. NADPH, IgG, GM14408, HIV-associated cancer)
- roman numerals (e.g. type II diabetes)
- Human gene symbols should be capitalized
- Type symbols should be capitalized (e.g. “type A”)
- Generally arabic > roman, except for established names (e.g. cranial nerve VII)
Uniqueness:
We strive for uniqueness. However, be careful when evaluating uniqueness. Various OMIMs that are apparently distinct have names that are the same, except with different lexicalizations.
Text definitions¶
We aim to write our own definitions following our standards, but for now we reuse roughly in order:
EFO > {Orphanet,NCIT} > DesignPattern > DOID
NCIT is generally favored over Orphanet, except for genetic non-cancer diseases.
We have overwitten some with our own. We aim for genus-differentia (but not in the style of DO which gets this wrong in many cases, for example, overstating the genus). More details on how to write simple, concise, and clear operational text definitions is here.
Synonyms¶
Use lowercase, even for initial letter, except for proper names (note: many synonyms remain with leading capitalization, this is improving).
Always annotate synonyms with xrefs. Many of these are currently DOID, Orphanet, GARD, etc IDs. We will add more directly referencing a publication (PMID CURIEs in the format PMID:XXXXXXX). Also add editor ID where appropriate (ORCID, in format with http://, for example: http://orcid.org/0000-0001-5208-3432).
Always indicate synonym scope (see below). Currently the following annotation properties are used:
- hasExactSynonym
- hasBroadSynonym
- hasNarrowSynonym
- hasRelatedSynonym
These are incorrect in many places where they have been brought in externally. Do not trust scope if there is no synonym xref other than DO.
Synonym scope:¶
Exact¶
The definition of the synonym is exactly the same as primary term definition. This is used when the same class can have more than one name.
For example, MONDO:0003321 hereditary Wilms' tumor and familial Wilms' tumor.
Narrow¶
The definition of the synonym is the same as the primary definition, but has additional qualifiers.
For example, MONDO:0004979 asthma and exercise-induced asthma
Broad¶
The primary definition accurately describes the synonym, but the definition of the synonym may encompass other structures as well. In some cases where a broad synonym is given, it will be a broad synonym for more than one ontology term.
For example, MONDO:0016264 autoimmune hepatitis and autoimmune liver disease
Related¶
This scope is applied when a word of phrase has been used synonymously with the primary term name in the literature, but the usage is not strictly correct. That is, the synonym in fact has a slightly different meaning than the primary term name. Since users may not be aware that the synonym was being used incorrectly when searching for a term, related synonyms are included.
For example, MONDO:0012996 AGAT deficiency has the related synonym disorder of glycine amidinotransferase activity.
We follow a lot of the same rules as Uberon for text mining: https://github.com/obophenotype/uberon/wiki/Using-uberon-for-text-mining
Synonym type:¶
EXCLUDE synonyms¶
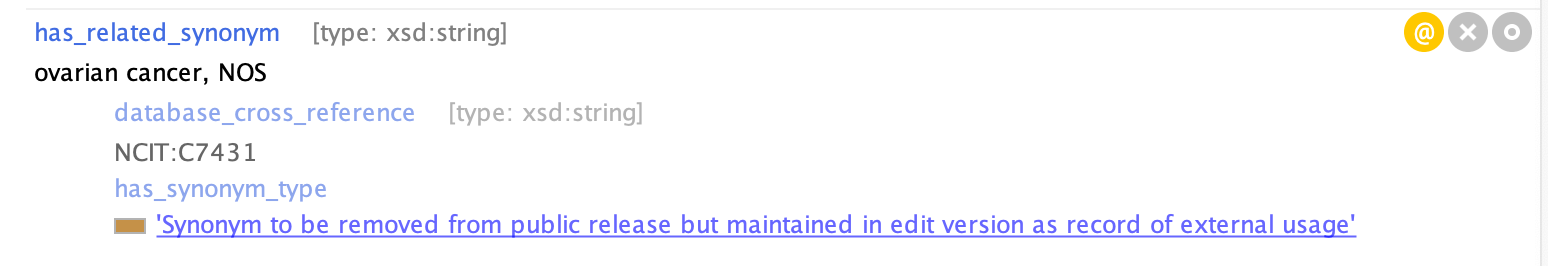
Some synonyms are annotated with EXCLUDE, e.g. “NOS” (not otherwise specified) synonyms. It is useful to have these in the edit version, but these are filtered on release.
For example, see MONDO_0002679 cerebral infarction or MONDO_0008170 'ovarian cancer'
DEPRACATE synonyms¶
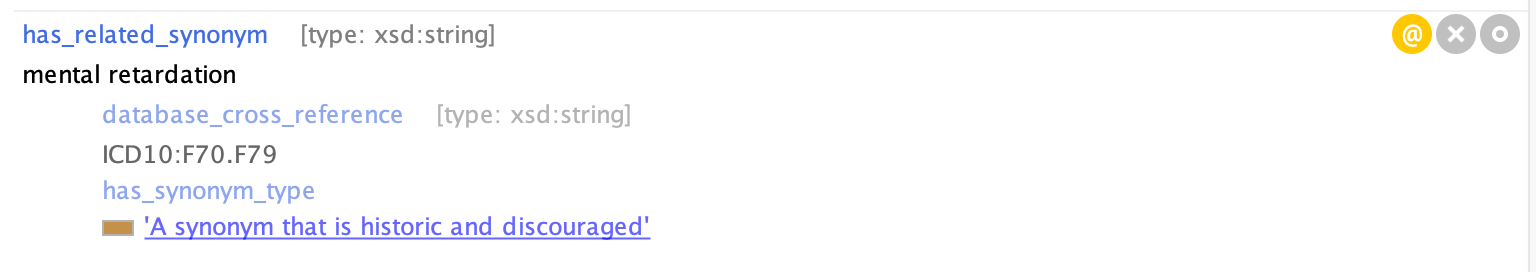
We may also mark synonyms with DEPRECATED. E.g. all occurrences of “mental retardation” should be “intellectual disability”
We try and avoid including things in this list: https://en.wikipedia.org/wiki/List_of_medical_eponyms_with_Nazi_associations but if it’s established (e.g. Wegener granulomatosis) may include as a synonym and mark DEPRECATED
For example, see MONDO_0001071 'intellectual disability'
How to add a synonym annotation in Protege¶
- Click on the synonym
- Click the @ symbol on the synonym
- Click
Annotations+ - Click
has_synonym_typeon the left - Click
Entity IRIon the top of the box - Click
Annotation propertieson the top of the box - Scroll to bottom, and show annotation below
synonym_type_property(click triangle) - Select appropriate synonym type (e.g.
A synonym that is historic and discouraged) - Click OK
Axiom Annotations¶
All axioms (logical and non-logical) SHOULD be annotated. We primarily annotate using the oio:source property. Synonyms and definitions are annotated using database_cross_reference (from oboInOwl) in the usual way.
The seed version of Mondo was created from external ontologies. We preserve both original axiom annotations where they are provided (e.g. DO) and we also stamp the originating ID.
Thus when we brought in
id: DOID:123 def: “blah” [http://foo.org]
We turn this into
id: MONDO:987 xref: DOID:123 {source=”Mondo:Equivalent”} def: “blah” [DOID:123, http://foo.org]
If we have two axioms that are identical except different annotations we merge into one with a union of annotations (we use owltools --merge-axiom-annotations)
Axiom Annotations on Xrefs¶
We care particularly about xrefs. We have two kinds of annotations on xrefs
- Semantic
- Provenance
We annotate these axioms to provide more specific semantics. Currently equivalence axioms ‘live’ in the xrefs, as it is easier to manage that equivalence axioms. See Protege screenshot below:
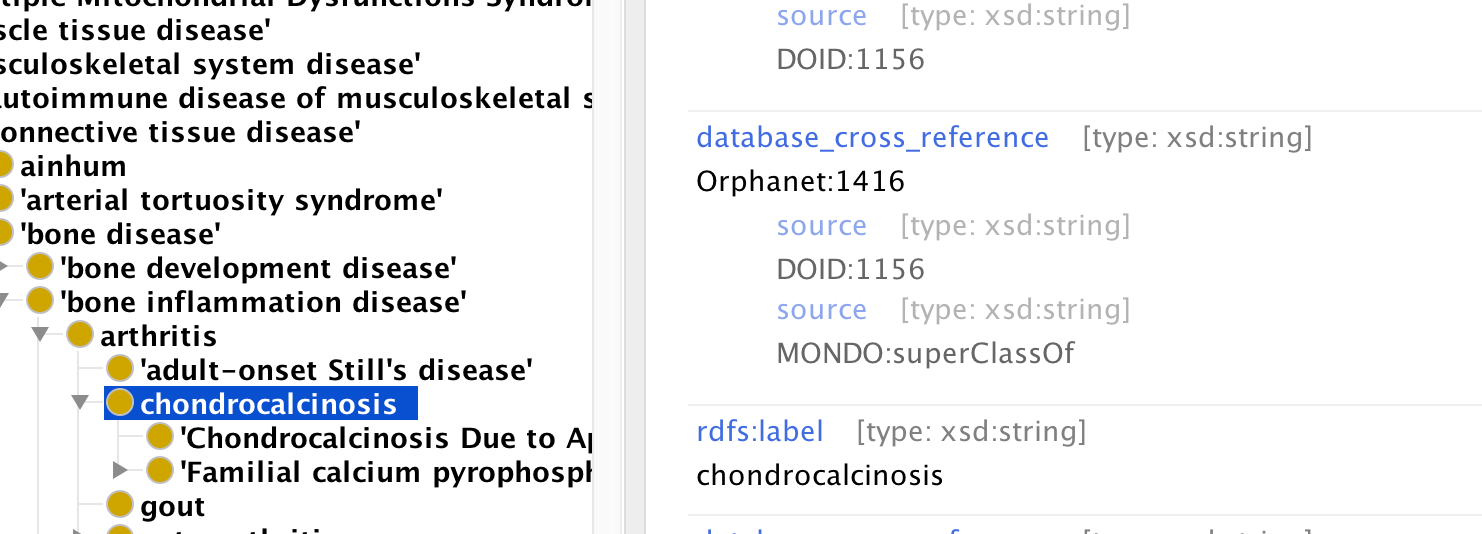
Here the xref between MONDO:chondrocalcinosis and Orphanet:1416 as two axiom annotations, one semantic and one provenance/consistency.
- We indicate the source of an xref using source, e.g. the source of the xref from our chondrocalcinosis class to Orphanet:416 is DOID:1156
- Note: we should consider changing the AP here. It’s not really the case that these are source any more, as they are added after the fact as an easy way to guage consistency of axioms across ontologies.
- We indicate the semantics of the xref using a source of one of
- MONDO:equivalentTo
- MONDO:superClassOf
- MONDO:subClassOf
- MONDO:OtherRelationship
- In the first 3 cases, these are interpreted strongly as an OWL axiom. Never use equivalentTo for ‘very close to’. Overuse will result in merges. See also docs on proxy merges
- MONDO:superClassOf and MONDO:subClassOf may be incomplete. The important one is equivalence, as they others can be inferred
- Note: we could of course use OWL axioms directly, but this is awkward for various reasons, so the logical axioms are maintained as annotated xrefs for now.
- We also use MONDO:obsoleteEquivalent and MONDO:equivalentObsolete for cases where we have exact 1:1 matches between an obsolete and a live class. We want to avoid making an equivalence axiom here.
Axiom Annotations on Logical Axioms¶
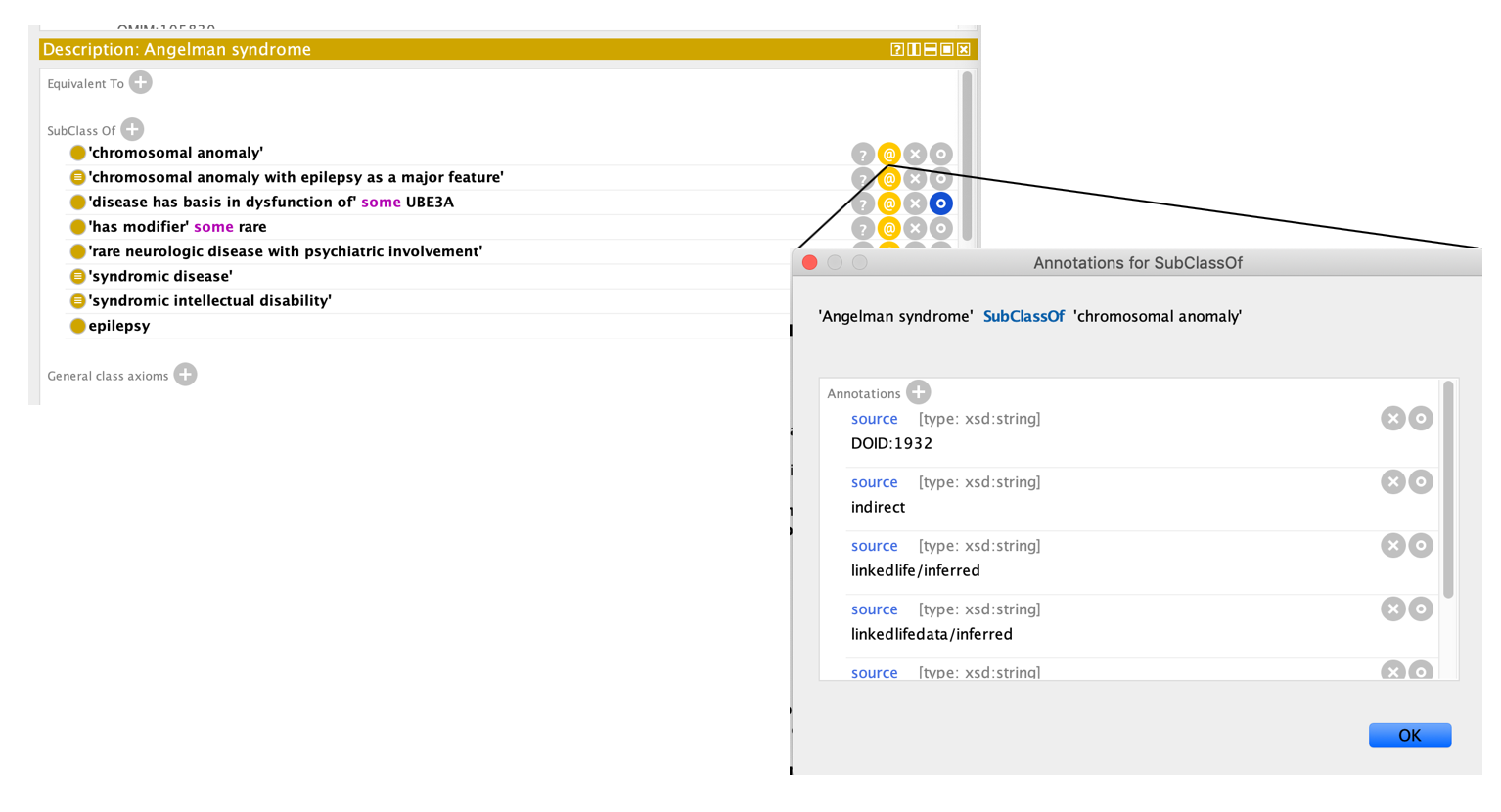
We aim to always state a reason why a subClassOf axiom exists. We currently use “oio:source” for everything (easier for roundtrip reasons). Currently subClassOf axioms may be annotated with
- List of IDs/CURIEs that support the axiom
- ONDO:Redundant, if it is entailed by other axioms
- Name of ontology editor
- PMIDs
- GitHub URLs
For example, see MONDO_0007113 'Angelman syndrome'
Axiom Annotations Summary Table¶
| Annotation | Description | What kind of axiom it can apply to | Editors only? | Example |
|---|---|---|---|---|
| MONDO:ambiguous | Used to indicate where there is a known case where this synonym is ambiguous with something else. | synonyms | N | MONDO:0009825 '5-oxoprolinase deficiency (disease)' (synonym: 5-oxoprolinase deficiency) |
| MONDO:design_pattern | If annotated on a synonym, the synonym was derived from a design pattern. | synonyms | N | MONDO:0009770 '3MC syndrome 1' (synonym: 3MC syndrome caused by mutation in MASP1) |
| MONDO:directSiblingOf | The term that is xref'd is a direct sibling of term. The goal was to capture where someone made an xref. | xrefs | N | MONDO:0008854 'Bardet-Biedl syndrome 1' (xref UMLS:C1858054) |
| MONDO:entailed | An inferred superclass (which is a redundant axiom) | subclassOf | Y | MONDO:0001594 'Achilles bursitis' |
| MONDO:equivalentObsolete | Used for cases where we have exact 1:1 matches between a live class in Mondo and an obsolete class in the source ontology.We want to avoid making an equivalence axiom (MONDO:equivalentTo) here. | xrefs | N | MONDO:0020499 'Nipah virus disease' |
| MONDO:equivalentTo | This is interpreted strongly as an OWL equivalence axiom. | xrefs | N | MONDO:0100087 'familial Alzheimer disease' |
| MONDO:kboom-pr-[number] | These are the probability scores from the kBoom algorithm. | xrefs | Y | MONDO:0008966 'Aagenaes syndrome' |
| MONDO:Lexical | Same as design_pattern. Should be replaced with specific design_pattern. | synonyms | N | MONDO:0010278 'Christianson syndrome' |
| MONDO:LexicalVariant | Similar to design_pattern, should be replaced with specific documentation about variant documentation. | synonyms | N | MONDO:0006018 'Wissler syndrome' |
| MONDO:notFoundInDiseaseSubset | This annotation is typically added to dbxefs from UMLS or NCIt, to indicate the term is not from the disease branch. | dbxef | N | MONDO_0015350 '17q11.2 microduplication syndrome' |
| MONDO:obsoleteEquivalent | Used for cases where we have exact 1:1 matches between an obsolete in Mondo and a live class in the source ontology. We want to avoid making an equivalence axiom (MONDO:equivalentTo) here. | xrefs | N | MONDO:0008858 'Behr syndrome' |
| MONDO:ontobio | Lexical matching method | subclassOf | N | MONDO:0012176 'Emanuel syndrome' |
| MONDO:patterns.... | A pattern was used to define the term or synonym, see: https://github.com/monarch-initiative/mondo/tree/master/src/patterns | definitions, synonyms | N | MONDO:0016593 'acquired ataxia' |
| MONDO:Redundant | An inferred superclass (which is a redundant axiom) | subclassOf | N | MONDO:0023543 'Katsantoni-Papadakou-Lagoyanni syndrome' |
| MONDO:relatedTo | Used when a term is not equivalent but similar. | xrefs | N | MONDO:0015350 '17q11.2 microduplication syndrome' |
| MONDO:subClassOf | This is interpreted strongly as an OWL subclass of axiom. | xrefs | N | MONDO:0017781 '12p12.1 microdeletion syndrome' |
| MONDO:superClassOf | This is interpreted strongly as an OWL superclass of axiom. | xrefs | N | MONDO:0017806 '15q overgrowth syndrome' |
Subsets¶
We use ORDO subClassOf as subset, thus we have subsets starting ordo_ for all their disease metaclassifications: group_of_disorders, etiological_subtype etc. We retain provenance as an axiom annotation.
We add our own subsets. These should be described in the header. They include subsets of classes that need further examination.
We use the n_of_one subset if a disease is described only in one individual or family. We try and retain these unless it is something truly lost in the midsts of time.